How to use - PPIR
Main menu:
How to added ptmPPIR in R
1. When in R click on "Packages" menu --> "Install package(s) from local zip files..."
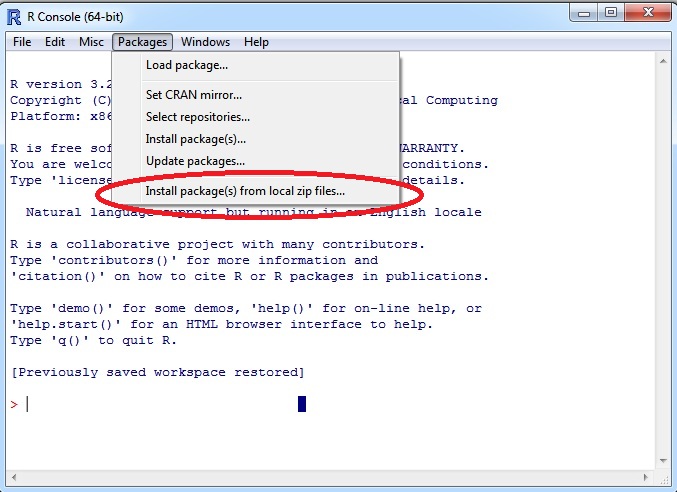
2. Browse for "PtmPPIR_0.5-1.zip" or "PtmPPIR_0.5-1.tar.gz" according to your OS
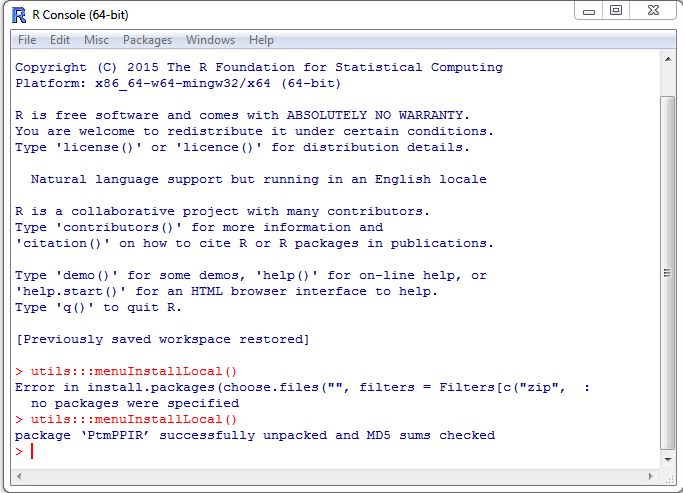
3. Wait untill PtmPPIR package is installed. If success, you should see message like following figure.
How to use ptmPPIR package
1. When in R type:
library(PtmPPIR)
2. R will then load PtmPPIR into its environment. You also see message "Loading required package: kernlab". This is because PtmPPIR require kernlab package for generating the models.
3. Basically, if you have sequence(s) in mind you can use a command as following:
- If only 1 sequence (must be 15 amino acids long, modifided site at the 8th position)type:predPPIR("CPKDEDYKQRTQKKA", ModType="acetylation")Please note that modtype here can be "acetylation", "ubiquitylation", and "phosphorylation"(case sensitive when typing)and you will see the result like:AAseq Res[1,] "CPKDEDYKQRTQKKA" "Out PPIRs"This indicates that the modified K (acetylated) is predicted as outside PPIRs by our model.
- If more than 1 sequence (also, must be 15 amino acids long, modifided site at the 8th position)
type:
predPPIR(c("CPKDEDYKQRTQKKA","ESKVFYLKMKGDYYR","RFGISREKQDTFALA","AKNSPLNKAEVHESY"),ModType="acetylation")
and you will see the result like:
AAseq Res
[1,] "CPKDEDYKQRTQKKA" "Out PPIRs"
[2,] "ESKVFYLKMKGDYYR" "In PPIRs"
[3,] "RFGISREKQDTFALA" "Out PPIRs"
[4,] "AKNSPLNKAEVHESY" "Out PPIRs"
This indicates that the modified K at 8th position of "ESKVFYLKMKGDYYR" is predicted as inside PPIRs by our model.
** It is more convenient if you provide the list of your sequences in the text file (one line per sequence)**
Then you can type:
myList = readLines("yourFile.txt")
then
predPPIR(myList, ModType="acetylation")
